Steve Vigdor, December 2, 2019
In our blog posts on evolution, we have shown that the rapid development of scientific techniques for mapping and understanding the genomes of humans and many other species has provided the strongest evidence we have of the central roles played by random mutations, natural selection and common descent in fueling the evolution of species. Genomic research has also provided strong evidence against the concepts of Intelligent Design, by demonstrating that biological features suggested by creationists to require supernatural design can, in fact, result naturally through many generations from random mutation, natural selection, the repurposing of genes originally developed to serve different functions, and changes to regulatory networks affecting gene expression. It is ironic, then, that logical next steps in genomic research have led in the 21st century to the development of technology that allows humans, in principle, to impose their own intelligence to alter the processes of natural (so-called Mendelian) inheritance and evolution.
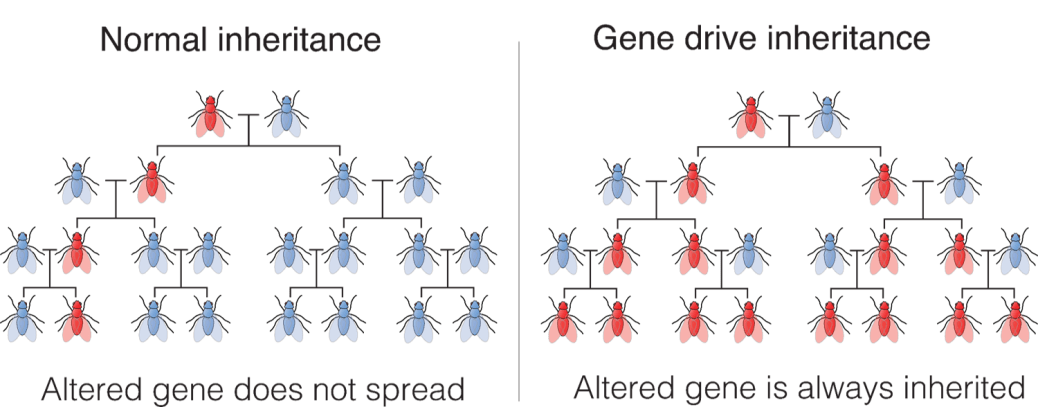
The new technology goes under the name of a gene drive. It provides a mechanism to strongly tilt the playing field in genetic inheritance, so that a genetic element edited by humans into a small percentage of individual organisms has a greatly enhanced probability to be passed from parent to offspring in sexual reproduction. Sexual reproduction involves the pairing of chromosomes contributed to the offspring from each parent. The matching chromosomes in each pair may contain different variants (called alleles) of the same gene, for example, a gene affecting eye or hair color. If one parent’s chromosomes contain two distinct alleles, the probability that the offspring will naturally inherit one of these alleles is 50%. However, gene drive technology can bias the inheritance probability of an engineered allele in one chromosome to nearly 100%.
As suggested in Fig. 1, the gene drive bias leads to a situation in which each successive generation sees a near-doubling of the frequency of finding the allele modified initially in a small fraction of individuals within a wild population. In a local population of rapidly reproducing species, the modified gene can affect the entire population after a modest number of generations. Thus, human selection of desired traits can dominate over natural selection. Whereas natural selection favors genomic modifications that lead over generations to improved fitness and fertility of species, human selection can lead to the opposite outcome. For example, a number of the prominently contemplated applications of gene drives would engineer sterility, or at least reduced fertility, into disease-bearing or pest populations, leading eventually to their extinction.
The interest of humans in creating organisms with more “desirable” traits is, of course, not new. Humans have long used selective breeding or cross-breeding of different breeds within a given species (e.g., dogs or horses) or hybridization of different animal or plant species to produce new organisms with greater robustness, or beauty, or utility to humans. But these efforts do not propagate through entire populations or species. Indeed, mating different species can lead to useful, but often sterile, offspring. An example is the mule, which is a sterile hybrid combining desired features of two parent species with different numbers of chromosomes: a male donkey and a female horse. More recently, humans have used genetic engineering to produce genetically modified crops, which may have increased robustness, but are not given any genetic advantage in passing their modified alleles along to offspring.
The new feature introduced by gene drives is the possibility for humans to modify entire populations and carry out ecosystem engineering. The possibilities for both good and harm are enormous. On the positive side, the interest in gene drives is driven by noble applications to the alteration or control of disease-carrying, invasive or endangered species populations. The technology holds the promise to save millions of lives. On the negative side are all the potential unintended consequences of altering ecosystems, inadvertently spreading genetic modifications to untargeted populations, or having desirable edited alleles corrupted by mutation to undermine the intended result. There are also possibilities of malevolent consequences, associated with weaponization of the technology to drive commercial, political or military advantages. Consequently, one must proceed with caution and thoughtful regulation in ongoing research and any eventual releases of gene drive organisms into wild populations.
In this post, we will describe in more detail the technology behind gene drives, some recent research results, their potential applications and dangers, the avenues of financial support and steps toward international regulation of ongoing research. We will also discuss erupting disagreements that mirror, in some ways, the tensions we have described on this site between scientific skepticism and science denial in other controversial topics at the interface between science and public policy.
1) Genetic Engineering
Genetic engineering refers to the use of biotechnology to remove, insert or replace small sections of DNA within a host organism’s cells, in order to alter the production of certain proteins or otherwise modify the host’s traits. It has been practiced as a discipline since 1972. The modified genes inserted or replaced have been taken from other organisms, including ones belonging to different species than the host, or more recently, artificially synthesized in the laboratory.
Early applications used recombinant DNA techniques, in which the desired gene is first chemically separated from its host DNA and inserted within a small, circular double-stranded DNA molecule called a plasmid. Plasmids are commonly found, physically separated from the chromosomal DNA, within bacterial cells, where they can independently replicate. Various techniques can then be used to combine copies of the modified plasmid with DNA samples from the targeted organism’s genome, and then to insert that combined DNA into target cells. Genetically modified organisms (GMOs) can then be regenerated from those modified cells, for example, from modified embryonic stem cells in animals.
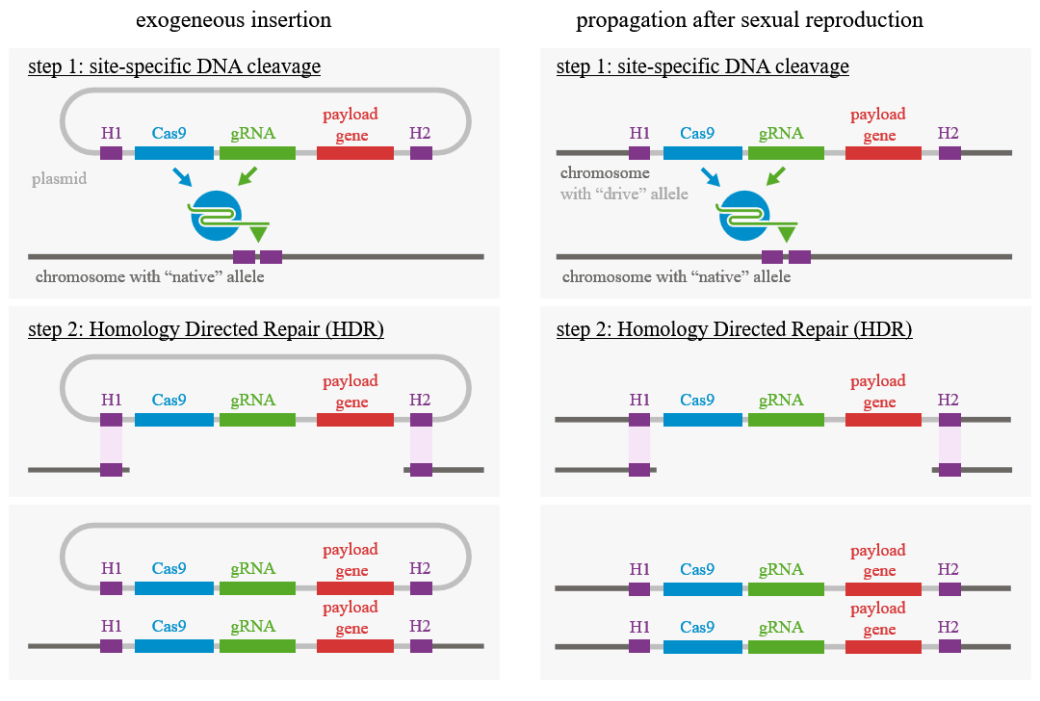
The new genetic material can be inserted into random or targeted locations within the host organism’s genome. But targeting is most efficiently carried out using the more modern techniques of genome editing. These techniques utilize artificially engineered enzymes, called nucleases, that are capable of cleaving the bonds at specific locations between adjacent nucleotides that comprise the elements of double-stranded DNA helices. The resulting breaks are then addressed by the cell’s natural DNA repair mechanisms. As indicated in Fig. 2, this may involve simply joining the broken ends without replacing the cleaved DNA section or, more frequently, copying a replacement from a template at the corresponding location in a similar (technically called “homologous”) or identical DNA molecule within the same cell, e.g., from the paired chromosome originally contributed by the other parent.

There are several techniques and nucleases used in gene editing, but the most commonly applied in recent years, since its development in 2012, is called CRISPR-Cas9. CRISPR stands for “clustered regularly interspaced short palindromic repeats.” That mouthful describes DNA sections similar to those found within the antiviral defense systems of certain bacteria. Cas9 is the “genetic scissors” protein paired with CRISPR. The CRISPR complex includes a synthetic guide RNA molecule that leads the Cas9 nuclease to cut the target DNA at the desired locations. The CRISPR complex can also include a “payload gene” that provides the template to be copied in homology-directed repair of the cut host DNA. CRISPR-Cas9 then provides a biomolecular analog to “find, cut and paste” operations in text editing. CRISPR-Cas9 has dramatically improved the efficiency and precision of genomic editing, and was consequently selected by Science magazine as the 2015 Breakthrough of the Year. It may well turn out to be the scientific breakthrough of the century. Jennifer Doudna and Emmanuelle Charpentier were awarded the 2020 Nobel Prize in Chemistry for their research demonstrating in detail the biomolecular composition of the CRISPR-Cas9 complex, and how bacteria use it to slice, dice and disable attacking viruses. As we will discuss in the next section, it is the technique most often associated with gene drives.
Genetic engineering techniques have been used to produce GMOs since the early 1970s. The first examples were intended for research use. The first commercial application, pioneered by Genentech in the late 1970s, was to the generation of insulin-producing bacteria. Genetically modified food has been sold since 1994. Most current GMO crops have been engineered to increase their resistance to insects, herbicides, diseases, environmental conditions and/or spoilage, and in some cases to improve their nutrient profile. By 2016, GMO techniques had been put to use over some 12% of global croplands.
A 2014 meta-analysis of GMO crop impacts found that where the technology had been adopted it had, on average, “reduced chemical pesticide use by 37%, increased crop yields by 22%, and increased farmer profits by 68%.” Despite a scientific consensus “that currently available food derived from GM crops poses no greater risk to human health than conventional food,” widespread public mistrust of the safety and possible unintended health consequences of genetically modified food have caused considerable controversy. Those safety concerns have led 38 countries, including 19 in Europe, but not the U.S., to officially prohibit GM crop cultivation. We will deal with the GMO food controversy in a future post on this site.
Another great hope for applications of genetic engineering is the use of gene therapy to cure genetic diseases, in addition to producing new hormones, vaccines and other drugs for the treatment of a wide variety of diseases. The aim of gene therapy is to replace or disrupt the functioning of genes responsible for diseases, either by correcting genetic mutations, or interfering with the expression of disease-causing genes, or inserting new genes that produce proteins that treat the disease.
Nearly all clinical trials to date have practiced somatic cell gene therapy, where the therapeutic genes are inserted into cells that affect only the biological functioning of the individual patient, and cannot be inherited by the patient’s offspring. Most of the trials focus on severe single-gene disorders, such as immuno-deficiencies, hemophilia, cystic fibrosis and sickle-cell anemia. Early clinical trials were often marked by failure or inconclusive benefits, but the last decade has seen a number of encouraging successes. The U.S. Food and Drug Administration has by now approved several gene therapy drugs for commercial sale.
Over the past few years, the application of gene editing techniques such as CRISPR have led to renewed hopes that gene therapy can cure some genetic diseases, viral diseases and cancer. A particular focus of current research is the use of CRISPR to disable DNA sections of viruses or bacteria that cause disease in humans. CRISPR complexes programmed to disable specific invading pathogens can reside within the helpful bacteria that are found in healthy human guts. Such a targeted approach has the potential, for example, to replace many of the more indiscriminate antibiotics currently in use, and to which many harmful bacteria are evolving resistance to pose a daunting worldwide medical threat for the coming decades. But all these approaches are still considered experimental and remain some years away from becoming medically approved practices.
A more controversial approach is so-called germline gene therapy, in which genes are edited directly within sperm or egg cells or embryos, so that the modifications may be passed on to future generations. The promise of removing disease-causing genetic defects not only in a patient, but in an entire line of descendants, is counterbalanced by concerns about unintended consequences and potential uses to enhance the inheritance of “desirable” traits not connected to diseases. The bioethical concerns have led a number of countries to prohibit germline gene therapy applications to humans, but the U.S. currently has no federal laws or regulations explicitly addressing human genetic modification.
Neither does China, which led to the worldwide 2018 controversy sparked by a claim from researcher Dr. He Jiankui that he had successfully used CRISPR-Cas9 to edit genes in embryos from seven couples, in order to disable a gene that forms a protein doorway allowing HIV – the virus that causes AIDS – to enter human cells. The edited embryos apparently resulted in one successful pregnancy, leading to the birth of genetically engineered twins. While the goal of engineering human resistance to HIV is laudable, He’s claim has not been published or independently confirmed, and has been judged wildly premature and irresponsible by most international scientists, given the still uncertain risks of germline editing.
The ethical issues have been addressed by several scientific and medical groups in the U.S. In 2017, a committee of the National Academy of Sciences and the National Academy of Medicine gave qualified support to genome editing in humans, once safety and efficiency of the techniques have been improved, “but only for serious conditions under stringent oversight.” Much earlier, the American Medical Association’s Council on Ethical and Judicial Affairs had concluded that “genetic interventions to enhance traits should be considered permissible only in severely restricted situations: (1) clear and meaningful benefits to the fetus or child; (2) no trade-off with other characteristics or traits; and (3) equal access to the genetic technology, irrespective of income or other socioeconomic characteristics.” As the techniques for genome editing continue to improve, and the suite of considered applications broadens, it will become imperative for the U.S. to adopt federal regulation guidelines and for international treaties restricting possible uses to be negotiated.
2) Gene Drive Concepts and Research
Not all genes are naturally subject to the rules of Mendelian inheritance in sexual reproduction. Evolutionary biologists have long known of the existence of so-called selfish genetic elements, which act by one mechanism or another to ensure their own overrepresentation in the genomes of offspring. They do this independently of natural selection, and regardless of whether their own reproduction is beneficial, neutral or harmful to the fitness and fertility of the species.
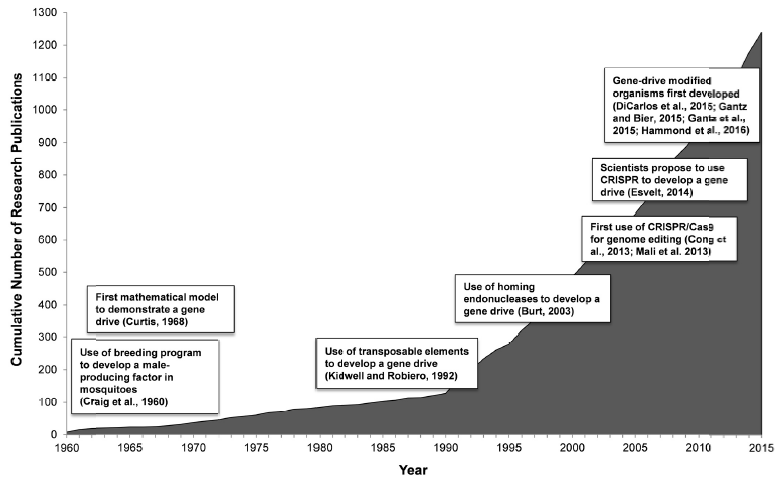
For example, it was found in the mid-20th century that some male mosquitoes of the Aedes aegypti species had a natural “male-producing factor” in the sex-determining section of their chromosomes, which ensured that most of their offspring would develop as males. In 1960, George Craig and colleagues then suggested that an environmental release of many such males could, over multiple generations, potentially “reduce the number of females below the [population] level required for efficient disease transmission.” Their concept was bolstered by subsequently developed mathematical models that demonstrated how such a selfish genetic trait could be driven to dominate an entire population. When such selfish genetic elements occur naturally, their exploitation to aid population control is an example of selective breeding, but its consideration began discussions about gene drives.
By the turn of the 21st century, as applications of genetic engineering grew more widespread, it was inevitable that scientists would begin to consider adapting some of the natural mechanisms used by selfish genetic elements to drive engineered genes to spread throughout selected populations. The first such suggestion was made by Kidwell and Ribeiro in 1992, who proposed using mobile sequences of DNA, known as transposable elements, to drive an engineered gene into a mosquito population. In 2003, Austin Burt proposed using a different natural selfish mechanism, centered around homing endonuclease genes. These are naturally occurring enzymes capable of recognizing (homing) a target DNA sequence in a homologous chromosome, cutting it, and then replacing the excised DNA section with a template from the endonuclease gene. In other words, they provide natural gene editing by a mechanism entirely similar to what later became possible with the development of CRISPR-Cas9 engineering.
It was the development of CRISPR-Cas9 that made engineered gene drives into a rapidly realizable technology, as first pointed out in 2014 by Kevin Esvelt and collaborators at the Wyss Institute for Biologically Inspired Engineering at Harvard University, one of the institutions where CRISPR-Cas9 gene editing technology was developed. The mechanism by which CRISPR-Cas9 gene drives ensure strong bias toward inheritance of the engineered gene is illustrated in Fig. 3. It basically uses insertion of an externally generated CRISPR complex into a target cell to engineer an edited homing endonuclease into one chromosome, which subsequently propagates itself into any natural chromosome with which it gets paired in sexual reproduction. Esvelt et al. pointed out that this technique “would represent an entirely new approach to ecological engineering,” but they also emphasized that “given the potential for gene drives to alter entire wild populations, and therefore ecosystems, the development of this technology must include robust safeguards and methods of control.”
As illustrated in Fig. 4, the seminal work of Kidwell and Ribeiro, of Burt, and of Esvelt, et al., have helped to spark an explosion of gene drive research over the past two decades. Within a year after the proposal by Esvelt, et al., four distinct proofs of principle for CRISPR-Cas9-generated gene drives were carried out in laboratory-based populations of yeast, fruit flies and two different strains of mosquitoes. The combination of these studies demonstrated how gene drives might be used to suppress an entire population of pest organisms, by editing in genes that reduce fertility, or to otherwise spread an engineered genome modification throughout a population.
A research paper published in September 2018 reported the first successful complete suppression of a population of caged organisms by gene drive modification. The modified organisms were male mosquitoes of the malaria-carrying species Anopheles gambiae. Half of the males in each sample population of 600 mosquitoes had one of their alleles associated with female development modified by CRISPR-Cas9, so that any female offspring they produced with two copies of the edited allele would be unable to lay eggs. After 7-11 generations, no further eggs were laid and the caged population reached extinction.
The extraordinarily rapid pace of research developments led the U.S. National Institutes of Health to ask the National Academies of Sciences, Engineering and Medicine in 2016 to convene a committee of experts charged with producing a study that would: “(1) review the state of the science and approaches to reduce unintended harms that could potentially result from developing and using gene-drive modified organisms; (2) discuss the ethical, legal, and social considerations attendant to field release of gene drives; and (3) determine the adequacy of existing governance mechanisms and risk assessment guidelines for the environmental and public health implications of using gene drives.”
In the resulting National Academies report, the committee notes: “The fast-moving nature of this field is both encouraging and a point of concern. Gene-drive modified organisms hold promise for addressing persistent or difficult-to-solve challenges, such as the eradication of vector-borne diseases and the conservation of threatened and endangered species. But the presumed efficiency of gene-drive modified organisms may lead to calls for their release in perceived crisis situations before there is adequate knowledge of ecological effects, and before mitigation plans for unintended harmful consequences are in place.” We will review some of the potential promised benefits and some of the possible harmful consequences in the ensuing sections.
But as though to emphasize the urgency of developing regulations regarding gene drive research and applications, two recent releases of genetically modified mosquitoes into wild populations have drawn considerable scrutiny. The more recent of these involved the first release of gene drive mosquitoes, engineered to drive sterility throughout the males in a population of malaria-carrying wild mosquitoes, into the wild in Burkina Faso in west Africa. Viewed as the first stage of a project to combat malaria in Africa, this release was of 10,000 already sterile males, which should not be able to pass on their edited genes, but which can be tracked to acquire data on the modified mosquitoes’ behavior and survival. But even this first stage has already drawn considerable criticism regarding the unforeseen consequences of ecosystem modification.
The earlier release of concern involved genetically modified, but not yet gene drive, mosquitoes. Nonetheless, the evolving results of this release give additional reasons to exercise caution in field studies of gene drive organisms. In this case, male mosquitoes of the Aedes aegypti species were genetically engineered to insert a dominant lethal allele, developed by the commercial company Oxitec Ltd., into a matched pair of their chromosomes, so that one copy of this allele would be guaranteed to pass on to offspring from mating with unmodified wild female mosquitoes. The engineered allele would lead to many fewer, and much weaker, offspring, which should be too weak to survive and breed themselves. The released mosquitoes’ genes were further edited to give them a fluorescent marker under certain light, so that they and their descendants could be distinguished and tracked after release into the wild. Approximately 450,000 of these genetically modified male mosquitoes were released each week over a period of 27 months in Jacobina, Brazil, a location surrounded in all directions by ecological conditions that suppress Aedes aegypti breeding.
The results anticipated from this release were a rapid reduction in the local Aedes aegypti population, with no leakage from the intruding gene pool into the surviving wild population. However, Yale researchers who have carried out genetic sampling of the Jacobina mosquito population during and after the release have found significant deviations from those expectations. First, they found that “the effectiveness of the release program began to break down after about 18 months, i.e., the population which had been greatly suppressed rebounded to nearly pre-release levels.” That could have occurred because the local female mosquitoes learned over time to avoid the genetically modified intruder males as mates.
The researchers also found that by the end of the release, portions of the genome of the intruder mosquitoes had been incorporated into the genome of the surviving population. These were not the engineered lethal and fluorescent genes, which did indeed gradually disappear from the population, but were distinct natural genes from the non-Brazilian populations that were used by Oxitec. Thus, the researchers concluded: “Evidently, rare viable hybrid offspring between the release strain and the Jacobina population are sufficiently robust to be able to reproduce in nature.”
It is not yet known whether the now hybrid Jacobina Aedes aegypti mosquito population might have attained improved robustness, or resistance to insecticides, or effectiveness as disease carriers, from the introduction of a foreign gene pool, or whether future mutations to their hybridized genomes might lead to such “improvements.” Such potential unintended consequences are among the outcomes that must be considered in deciding whether to release gene drive organisms. The National Academies study summarized the questions for gene drive releases this way:
“Will applications of gene drives be safe? Will they be effective? Will they have unintended consequences for the environment or public health? Do we know enough to release gene-drive modified organisms into the wild? Is using a gene drive to suppress or eliminate a pest species a good idea? What can scientists do to reduce risks to humans, other organisms, and the environment? How do we decide where gene-drive modified organisms might get released? What should governments do? Who gets to decide?”
3) Applications of Gene Drives
The National Academies report tabulates the potential applications of gene drives currently under discussion according to the following classification.
Public Health:
- Control or alter organisms that carry infectious diseases that affect humans, such as dengue, malaria, Chagas and Lyme disease
- Control or alter organisms that directly cause infection or disease, such as Schistosomiasis
- Control or alter organisms that serve as reservoirs of disease, such as bats and rodents
Ecosystem Conservation:
- Control or alter organisms that carry infectious diseases that threaten the survival of other species
- Eliminate invasive species that threaten native ecosystems and biodiversity
- Alter organisms that are threatened or endangered
Agriculture:
- Control or alter organisms that damage crops or carry crop diseases
- Eliminate weedy plants that compete with cultivated crops, e.g., by reversing their development of herbicide resistance
Basic research:
- Alter model organisms to carry out research on gene drive function and effects, species biology, and mechanisms of disease
Much of the ongoing research and field tests are aimed specifically at stopping the spread of malaria, a disease that has been persistently resistant to effective treatment. Malaria still kills about half a million people every year worldwide, predominantly children under five years of age, who are the most vulnerable. The Bill and Melinda Gates Foundation has provided substantial funding to these efforts, helping to support both Oxitec Ltd. (the company behind the genetically modified mosquito release in Brazil) and Target Malaria, a non-profit organization behind the Burkina Faso release. Target Malaria specifically aims at reducing the population of malaria-transmitting mosquitoes in sub-Saharan Africa. As they point out on their website: “While there are more than 3,500 species of mosquito worldwide and 837 in Africa, only three 3 very closely related species are responsible for most transmission of the disease: Anopheles gambiae, Anopheles coluzzii and Anopheles arabiensis. Our [gene drive] technology specifically targets these Anopheles species and so should not affect other types of mosquitoes or insects in the surrounding environment.”
A broader set of targets for gene drive technology has been set by the government of New Zealand, which announced in July 2016 an ambitious Predator Free 2050 program to rid the entire country of all its invasive vertebrate predator species that have threatened the islands’ natural biodiversity, driving many native species to extinction, and caused extensive agricultural losses. The New Zealand government and various philanthropic groups have pledged to provide about NZ$3 billion by 2050 to the cause of eradicating rats, brushtail possums, stoats and other predator species. Part of the envisioned program involves traditional methods using traps and poisons. James Russell, the leader of research and development for Predator Free 2050, has said: “We’re in a relatively unique position in New Zealand, where people are really, really willing to kill for conservation. It’s kind of a national pastime.”
But it is recognized that traps and poisons will not be nearly enough to realize New Zealand’s ambitious goals. Thus, an important part of the New Zealand project is the development of CRISPR-Cas9 gene drives that can cause disruption throughout the targeted populations of genes vital for survival or reproduction, or that can make the animals more susceptible to selected poisons. An analogous project being pursued by a coalition of seven worldwide organizations is called GBIRd, aiming at Genetic Biocontrol of Invasive Rodents on islands around the globe. One particular approach under development in New Zealand, known as the Trojan Female Technique, involves exploiting either natural mutations or gene drives that would be passed down among females, without affecting their own fertility, but would lead them to produce sterile male offspring, whose sperm have deteriorated swimming capability. There are plans as well in New Zealand for a widespread education campaign that will attempt to overcome the anticipated public resistance to the wide dispersal of genetically modified animals into the wild.
One should not, however, overlook the application of gene drive technology to basic research about its effectiveness and limitations. Surprisingly, the largest single source of funding for this type of research is being provided by the U.S. Defense Advanced Research Projects Agency (DARPA). In July 2017, DARPA announced the award of $65 million in grants to seven research teams collaborating on a Safe Genes program. DARPA explains its interest in the technology this way: “The science of gene editing, including gene drive technology, has been advancing at a rapid pace in the laboratory. These leaps forward in potential capability, however, have not been matched by advances in the biosafety and biosecurity tools needed to protect against potential harm if such technologies were accidentally or intentionally misused, nor does data exist on how such technologies would actually function in the far more complex real world.”
Among the research topics for the Safe Genes program are the development of techniques to switch gene drives on and off, to limit their geographical spread, to monitor their impacts both within and beyond the targeted species populations, and to limit or reverse their potential threats to biosafety and biosecurity. The scope of the funded program makes it clear that DARPA interprets gene drives as posing significant threats. That brings us in the next section to consider the potential dangers of gene drives in more detail. In the final section of this post, we will briefly describe some of the countermeasure technologies being pursued under the Safe Genes program, along with issues regarding the development of global guidelines and possible regulations governing gene drive research and applications.
4) Potential Dangers of Gene Drives
Many people are likely to object to the application of gene drives on ethical or religious grounds, as the technology offers humans the possibility to “play God.” But in this section, we are interested in exploring what could go wrong technically. We will describe several general areas of risk:
- The unintended dispersal of gene drives beyond the targeted population, or even the targeted species;
- Ripple effects on entire ecosystems to which the targeted population belongs;
- Natural evolutionary pressures that may neutralize, undermine or corrupt the intended gene drive effect;
- Applications of gene drive technology to “purify” species that are neither threats nor under threat themselves, by enhancing human-desired traits;
- Weaponization of gene drives as biological agents to gain military or agribusiness advantages.
What if a gene drive intended to eradicate a local pest population spreads beyond that population? Various mechanisms for such an unintended spread have been identified. A genetically modified specimen may migrate itself, or its germ cells may migrate, to affect other populations via processes known as gene flow. Migration can occur even for land animals allegedly confined to isolated islands. A famous example occurred during invasive species studies in New Zealand, when one among many radio-collared rats released on an initially rat-free offshore island managed to evade capture, swim to a nearby island, and avoid being trapped there for 18 weeks. Furthermore, migration of organisms is often assisted, either intentionally or unwittingly, by human transportation, potentially over quite long distances.
Many plants and some marine invertebrates disperse their genome by the movement of germ cells, rather than of entire organisms. The most familiar example is wind-borne or insect-facilitated pollen dispersal, which has the potential to move engineered genes over long distances into distinct populations or even different plant species, where it can cause unintended hybridization. It is also well known that a significant factor in natural evolution is horizontal gene transfer, where several distinct mechanisms other than mating can cause transfer of DNA segments between organisms belonging to two distinct species. For example, viruses exchanged between species can effect such a transfer.
Gene flow and horizontal gene transfer can potentially harm non-targeted populations or species, and that can happen in communities that have not been informed or consulted about the gene drive risks. That potential exists even if the gene drive is intended to benefit, rather than eradicate, the target population, since the engineered genes can be maladapted to the environment and ecosystem in which the non-targeted population exists. Such maladaptation can potentially be undone by natural selection within the non-targeted population, but only if the genetic intrusion occurs sufficiently slowly.
One must also consider the non-trivial ecological consequences of rapidly changing the abundance of a gene-drive-targeted species. How does this affect the local competition among species and the local food chain? How would elimination of one population affect the survival of other species that may have co-evolved to prey specifically on the eliminated species? Even mosquitoes, the most widely considered gene drive targets, serve as food for various aquatic predators and for bats. Some might consider bees to be pests suited for gene drive population reduction, but what species would replace their critical role as pollinators for 70 of the hundred or so crop species that feed 90% of humans around the world? Might the elimination of one pest species lead to increased robustness of another? Might a gene drive intended to limit mosquitoes’ ability to transmit one disease lead to their increased susceptibility as hosts for another, equally dangerous virus? Might a gene drive to suppress a non-native weed population lead to the loss of habitat for native species, or even the establishment of a second, more resilient invasive species? These are among risks that will need to be evaluated carefully in research to be carried out for each contemplated gene drive application.
Natural selection has, in the past, often overcome humans’ attempt to control or eradicate certain species. A particularly prominent example is the development of antibiotic resistance in bacterial species. What is the chance that targeted populations will evolve resistance to applied gene drives, possibly growing more robust or even more harmful in the process? As the National Academies report on gene drives points out, “the lower the equilibrium population mean fitness becomes after the introduction of a gene drive, the stronger the selection pressure will be on any beneficial resistance mutant that arises even though the rate of these mutations will be lower as well.” A comparable growth in selection pressure is known to occur naturally in the development of effective antibodies to fight off viral or bacterial infections, after weakly effective antibodies may have had some initial success in reducing the abundance of the invading antigens.
Opponents of gene drive research are especially concerned about the powerful temptations to humans to use gene drives to “enhance” conservation, purify species, or attain military or business advantages. They worry, for example, that commercially patented gene drives can be used to speed up the introduction of genetically modified traits into seed harvests before the unintended consequences are understood, or released into wild weed species to make them more susceptible to a particular proprietary herbicide, such as Roundup. They point out possible military uses of gene drives as biological agents to spread disease or toxins, or to attack an enemy country’s food production. They find the extensive DARPA financial support for gene drive research to be ominous, although DARPA claims it is driven by Department of Defense concerns about biosafety and biosecurity that are similar to the concerns of gene drive opponents.
The opponents also worry about attempts at artificial enhancement of conservation, such as the GBIRd project, going rogue and producing “purified” species. Such possibilities need to be taken seriously, in light of the long history, described elsewhere on this site, of eugenics arguments calling for sterilization of human populations that some groups view as “inferior.”
In short, both the promise and the risks of gene drive technology are too serious to be overlooked. The need for guidance and regulations governing the responsible pursuit of research on this topic is urgent. And there needs to be international agreement on such guidance since, as the National Academies group has pointed out: “After release into the environment, a gene drive knows no political boundaries.”
5) International Guidelines and Controversy
International regulations governing the development and use of GMOs is provided, in principle, by the United Nations Convention on Biological Diversity (CBD), as implemented through the Cartagena and Nagoya Protocols. The Cartagena Protocol was drafted in 2000 and has been ratified to date by 171 countries, not including the U.S. Many countries are developing their own regulatory systems for GMOs and gene drives under the Cartagena Protocol. But the U.S. does not currently have a clear policy for collaborating with other countries, or for sharing best practices and concepts. Within the U.S. there is a Coordinated Framework for the Regulation of Biotechnology, but the federal agencies (FDA, EPA, USDA) included in the Framework do not have clear lines of authority or responsibility for gene drive research.
The governing body of the U.N. Convention on Biological Diversity, known as the Conference of the Parties, meets every two years to sort through thorny international issues, such as guidelines for gene drive research. Their most recent meeting was in November 2018 in Sharm-el-Sheikh, Egypt. The year leading up to that meeting was marked by an intense controversy, sparked especially by opponents of gene drive research, who lobbied for a global moratorium on further research.
The opponents attempted to stir up a scandal analogous to the Climategate scandal, in which e-mail messages hacked from researchers at the University of East Anglia in the U.K. were interpreted to suggest that climate scientists were conspiring to doctor data and suppress publication of alternative views. In the gene drive case, e-mails were obtained under a Freedom of Information Act request from the account of Todd Kuiken, a synthetic biology researcher at North Carolina State University in Raleigh, and a member of the CBD’s Ad Hoc Technical Expert Group (AHTEG) on Synthetic Biology. The e-mails do not reveal any nefarious activity, but they do reveal coordination of efforts among proponents of ongoing, but carefully regulated, gene drive research, and they discuss some of the Safe Genes program research being funded by DARPA.
In particular, a public relations firm, Emerging Ag – working on behalf of its non-profit client Target Malaria and funded in part by the Bill and Melinda Gates Foundation to “increase awareness, understanding, and acceptance of possible gene drive applications for public good purposes” – was coordinating with Kuiken and other AHTEG members to recruit scientists to participate in an open online forum on synthetic biology, intended to inform the CBD. Given that the online forum is open to anyone, and that many civil society organizations had already banded together to sign a public call for a moratorium on gene drive research, efforts to get the researchers involved to take more active part in the conversation appear to be quite natural.
Nonetheless, SynBioWatch, a collaboration of several non-governmental organizations that monitors “next-generation genetic engineering,” posted the full set of pretty mundane e-mails, under the title Gene Drive Files, on their website, headed by the following sensationalized, fear-inducing claims:
“The Gene Drive Files are a trove of emails and other records uncovered by civil society investigators. The Files reveal the US military as the number one funder and influence accelerating the development of “gene drives”, a controversial and powerful new genetic extinction technology. The files also reveal that a previously undisclosed gene drive “advocacy coalition,” was run by a private PR firm who received $1.6 million in funds from the Bill and Melinda Gates Foundation. They appear to have used covert lobbying tactics to influence expert UN discussion.”
Although, as outlined in the preceding section, the risks of gene drives released into the wild are real, and also are universally acknowledged by researchers in the field, it would be misguided to suspend further research. The ethics of eliminating research on technology that is already out there, and that has the potential to save millions of lives, are questionable. But even more importantly, the thrust of much of the ongoing research, and in particular of that funded by DARPA, is on finding scientific ways to mitigate the risks.
Kevin Esvelt, the young researcher whose 2014 paper opened the CRISPR gene drive floodgates, is strongly concerned about the risks and is leading one of those DARPA-funded research projects. In the wake of the rush to develop gene drives, Esvelt has said: “We are walking forwards blind. We are opening boxes without thinking about consequences. We are going to fall off the tightrope and lose the trust of public.” He and his collaborators are now working on a significant tweak to the technology, known as daisy-chain gene drives, intended to limit the spread of gene drives beyond the locally targeted population.
The concept of one form of the daisy chain is illustrated in Fig. 5. In contrast to a normal gene drive, where a single CRISPR-Cas9 implant includes all the elements needed to copy itself into a homologous chromosome, and thereby to ensure its own inheritance, those elements are separated and inserted into different sections of the genome in a daisy chain drive. For example, the guide RNA that instructs the Cas9 nuclease where to cut can itself be inserted into a distinct part of the genome, where it does not replicate itself, and thus does not bias its own inheritance in subsequent generations. The remainder of the CRISPR complex does get replicated in homologous chromosomes during sexual reproduction, but only when the guide RNA is present to launch the operation. But the guide RNA will appear in only half the offspring of mating between the originally engineered and wild specimens, and may get reduced further in frequency in subsequent generations by natural selection.

The example illustrated in Fig. 5 contains an additional possible engineering trick to penalize the mating of daisy drive descendants with wild specimens, by also swapping two essential genes between two distinct chromosomes in the engineered specimens. When a daisy drive descendant lacking the guide RNA mates with a wild specimen, half of the offspring will be born without one or the other of these two essential genes, and will not survive. This penalty limits both the generational longevity of the engineered genes and their spread to populations beyond the one originally targeted.
Yet other ongoing research focuses on the development of a chemical “on-off” switch for gene drive operation, which could allow control of the inheritance bias in case of undesirable consequences. For example, a British team has developed in the laboratory a variant of the Cas9 enzyme whose expression is dependent on the presence of a distinct amino acid known as BOC. The gene drive will then only work if the target organisms ingest BOC, and scientists could regulate the gene drive in the wild by adding or withdrawing BOC from the local environment. Similar research is being pursued at the Broad Institute of MIT and Harvard, as part of DARPA’s Safe Genes program.
It is certainly prudent to delay any wild release of gene drive organisms until such research may develop approaches that offer greater control over the implementation. Such prudence is consistent with both the recommendations of the 2016 National Academies study and the guidelines set forth by the CBD at their 2018 meeting. After debating the contentious issues over two weeks in Sharm-el-Sheikh, the CBD’s Conference of the Parties worked out a compromise that allows research to proceed cautiously. As summarized in a Washington Post article, “Research efforts would have to show that the re-engineered organisms pose no hazard. Any field test would require the ‘free, prior and informed consent’ of people who live in the areas affected by the experiment.”
The caution is appreciated both by researchers in the field and by organizations that stand to benefit strongly from the technology. Kevin Esvelt, for example, has said “he would oppose any for-profit use of the technology. He says gene drives are more likely to be embraced by the public — and thus more likely to save lives — if people don’t fear that profits rather than public health is the major motivation.” Fredros Okumu, director of science at the Ifakara Health Institute in Tanzania, where malaria poses an enormous threat, says: “There’s nothing we have that carries zero risk. The question is, how do we mitigate those risks so that we can maximize the gains associated with, potentially, the huge value that can come out of gene-drive research?”
The issue of international guidelines for both research and field tests is bound to come up again at the 2020 meeting of the CBD. And in the U.S., where Senate opposition prevented ratification of the U.N. Convention on Biodiversity, clear lines of authority and regulatory guidelines for approval of gene drive research and applications are urgently needed.
References:
https://en.wikipedia.org/wiki/Gene_drive
https://en.wikipedia.org/wiki/Selective_breeding
https://en.wikipedia.org/wiki/Crossbreed
https://en.wikipedia.org/wiki/Hybrid_(biology)#Anthropogenic_hybridization
https://en.wikipedia.org/wiki/Genetic_engineering
https://en.wikipedia.org/wiki/Synthetic_biology
https://en.wikipedia.org/wiki/Recombinant_DNA
https://en.wikipedia.org/wiki/Genetically_modified_organism
https://en.wikipedia.org/wiki/Genetically_modified_crops
https://en.wikipedia.org/wiki/Genetically_modified_food_controversies
https://en.wikipedia.org/wiki/Nuclease
https://en.wikipedia.org/wiki/CRISPR_gene_editing
https://en.wikipedia.org/wiki/Gene_therapy
W. Klümper and M. Qaim, A Meta-Analysis of the Impacts of Genetically Modified Crops, PLoS One 2014, e111629. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4218791/
K. Sheikh, Is Crispr the Next Antibiotic?, New York Times, October 28, 2019.
https://www.nytimes.com/2019/10/28/health/crispr-genetics-antibiotic-resistance.html?auth=login-email&emc=edit_th_191105%3Fcampaign_id%3D2&instance_id=13527&login=email&nl=todaysheadlines®i_id=353363561105&segment_id=18515&user_id=c1558c49a4e3cd3e0fd628a032cecc8e
https://www.cdc.gov/drugresistance/index.html
M. Marchione, Chinese researcher claims first gene-edited babies, Associated Press, Nov. 26, 2018. https://www.apnews.com/4997bb7aa36c45449b488e19ac83e86d
R. Stein, Facing Backlash, Chinese Scientist Defends Gene-Editing Research On Babies, National Public Radio, Nov. 28, 2018
https://www.npr.org/sections/health-shots/2018/11/28/671375070/facing-backlash-chinese-scientist-defends-gene-editing-research-on-babies
Human Genome Editing: Science, Ethics and Governance, http://nationalacademies.org/cs/groups/genesite/documents/webpage/gene_177260.pdf
The Council on Ethical and Judicial Affairs, American Medical Association, Ethical Issues Related to Prenatal Genetic Testing. https://www.ncbi.nlm.nih.gov/pubmed/7921302
https://en.wikipedia.org/wiki/Selfish_genetic_element
Craig, G.B., Jr., W.A. Hickey, and R.C. Vandehey, An Inherited Male-Producing Factor in Aedes aegypti. Science 132, 1887 (1960). https://science.sciencemag.org/content/132/3443/1887
Curtis, C.F., Possible Use of Translocations to Fix Desirable Genes in Insect Pest Populations. Nature 218, 368 (1968). https://www.nature.com/articles/218368a0
Kidwell, M.G., and J.M. Ribeiro. Can Transposable Elements Be Used to Drive Disease Refractoriness Genes into Vector Populations? Parasitology Today 8, 325 (1992). https://www.sciencedirect.com/science/article/pii/016947589290065A
Burt, A., Site-Specific Selfish Genes as Tools for the Control and Genetic Engineering of Natural Populations. Proc. Biol. Soc. 270, 921 (2003). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1691325/
https://en.wikipedia.org/wiki/Homing_endonuclease
Esvelt, K.M., A.L. Smidler, F. Catteruccia, and G.M. Church, Concerning RNA-Guided Gene Drives for the Alteration of Wild Populations. eLife 3, e03401 (2014). https://elifesciences.org/articles/03401
DiCarlo, J.E., A. Chavez, S.L. Dietz, K.M. Esvelt, and G.M. Church. Safeguarding CRISPR-Cas9 Gene Drives in Yeast. Nat. Biotech. 33, 1250 (2015). http://arep.med.harvard.edu/pdf/DiCarlo_nbt_2015.pdf
Gantz, V.M., and E. Bier, The Mutagenic Chain Reaction: A Method for Converting
Heterozygous to Homozygous Mutations. Science 348, 442 (2015). https://science.sciencemag.org/content/348/6233/442.full
Gantz, V.M., N. Jasinskiene, O. Tatarenkova, A. Fazekas, V.M. Macias, E. Bier, and A.A. James, Highly Efficient Cas9-Mediated Gene Drive for Population Modification of the Malaria Vector Mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. 112, E6736 (2015). https://www.pnas.org/content/112/49/E6736
Hammond, A., R. Galizi, K. Kyrou, A. Simoni, C. Siniscalchi, D. Katsanos, M. Gribble, D. Baker, E. Marois, S. Russell, A. Burt, N. Windbichler, A. Crisanti, and T. Nolan, A CRISPR-Cas9 Gene Drive System Targeting Female Reproduction in the Malaria Mosquito Vector Anopheles gambiae. Nature Biotechnology 34, 78 (2016). https://www.nature.com/articles/nbt.3439
K. Kyrou, A.M. Hammond, R. Galizi, N. Kranjc, A. Burt, A.K. Beaghton, T. Nolan and A. Crisanti, A CRISPR-Cas9 Gene Drive Targeting Doublesex Causes Complete Population Suppression in Caged Anopheles gambiae Mosquitoes, Nature Biotechnology 36, 1062 (2018). https://www.nature.com/articles/nbt.4245
Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values (National Academies Press, 2016), http://nap.edu/23405
https://www.theguardian.com/world/2018/nov/25/gm-mosquitoes-released-burkina-faso-malaria-gene-drive
https://www.pri.org/stories/2019-07-04/burkina-faso-fighting-malaria-genetically-modified-mosquitoes
https://targetmalaria.org/our-work/
B.R. Evans, et al., Transgenic Aedes aegypti Mosquitoes Transfer Genes into a Natural Population, Scientific Reports 9, 13047 (2019), https://www.nature.com/articles/s41598-019-49660-6#Abs1
https://newatlas.com/science/genetic-engineering-mosquito-experiment-goes-wrong/
https://newatlas.com/science/gm-mosquito-study-oxitec-rebuttal/
https://en.wikipedia.org/wiki/Predator_Free_2050
B. Owens, Behind New Zealand’s Wild Plan to Purge All Pests, Nature 541, 148 (2017). https://www.nature.com/news/behind-new-zealand-s-wild-plan-to-purge-all-pests-1.21272
http://www.scoop.co.nz/stories/SC1701/S00024/predator-free-nz-expert-qa.htm
https://www.geneticbiocontrol.org/
https://www.darpa.mil/program/safe-genes
https://gizmodo.com/why-darpa-is-investing-big-in-gene-drives-1821028638
https://en.wikipedia.org/wiki/Gene_flow
J.C. Russell, D.R. Towns, S.H. Anderson and M.N. Clout, Intercepting the First Rat Ashore, Nature 437, 1107 (2005). https://www.nature.com/articles/4371107a
https://en.wikipedia.org/wiki/Horizontal_gene_transfer
https://www.bbc.com/future/article/20140502-what-if-bees-went-extinct
Frequently Asked Questions on the Cartagena Protocol on Biosafety, https://2001-2009.state.gov/g/oes/rls/or/2004/29751.htm
Reckless Driving: Gene drives and the end of nature, http://www.synbiowatch.org/2016/08/reckless-driving/
https://en.wikipedia.org/wiki/Coordinated_Framework_for_Regulation_of_Biotechnology
http://www.synbiowatch.org/gene-drives/
http://www.synbiowatch.org/wp-content/uploads/2016/09/ETC_genedrivers_v10.pdf
http://genedrivefiles.synbiowatch.org/2017/12/01/gates_foundation_pr/
The Case for a Global Moratorium on Genetically-Engineered Gene Drives,
http://www.etcgroup.org/sites/www.etcgroup.org/files/files/cbd_cop_13_gene_drive_moratorium_briefing.pdf
Drive Safely: Scientists Must Persist in Pointing Out the Environmental Dangers of Gene Editing, Nature editorial, December 7, 2017, https://www.nature.com/magazine-assets/d41586-017-08214-4/d41586-017-08214-4.pdf
K.V. Brown, This Scientist is Trying to Stop a Lab-Created Global Disaster, Splinter News, June 27, 2016. https://splinternews.com/this-scientist-is-trying-to-stop-a-lab-created-global-d-1793857858
http://www.sculptingevolution.org/genedrives
http://www.sculptingevolution.org/daisydrives
https://www.responsivescience.org/pub/daisydrives
https://allianceforscience.cornell.edu/blog/2018/07/new-gene-drive-off-switch-assuage-fears-critics/
T. Suzuki, et al., Switchable Genome Editing via Genetic Code Expansion, Scientific Reports 8, 10051 (2018), https://www.nature.com/articles/s41598-018-28178-3.pdf
https://www.broadinstitute.org/news/researchers-identify-molecules-rein-crispr-systems
https://singularityhub.com/2019/01/02/gene-drives-didnt-get-banned-last-year-so-whats-next-for-them/
